Liyana Shatar and Fariza Hanim Suhailin*
*Corresponding author: farizahanim@utm.my
Surface-Enhanced Raman Spectroscopy (SERS) has emerged as a sensitive and label-free technique for detecting biomolecules in clinical, environmental, and food safety applications (Cialla-May et al., 2024). The performance of a SERS biosensor depends significantly on its biorecognition element, which selectively binds the target analyte and enables effective signal enhancement. Among the various recognition elements available, aptamers are gaining popularity due to their high binding affinity, structural versatility, and synthetic accessibility (Sequeira-Antunes & Ferreira, 2023). Unlike antibodies and DNA probes, aptamers are smaller in molecular weight (10–30 kDa), more chemically stable, and can be produced synthetically at lower cost (Muhammad & Huang, 2021).
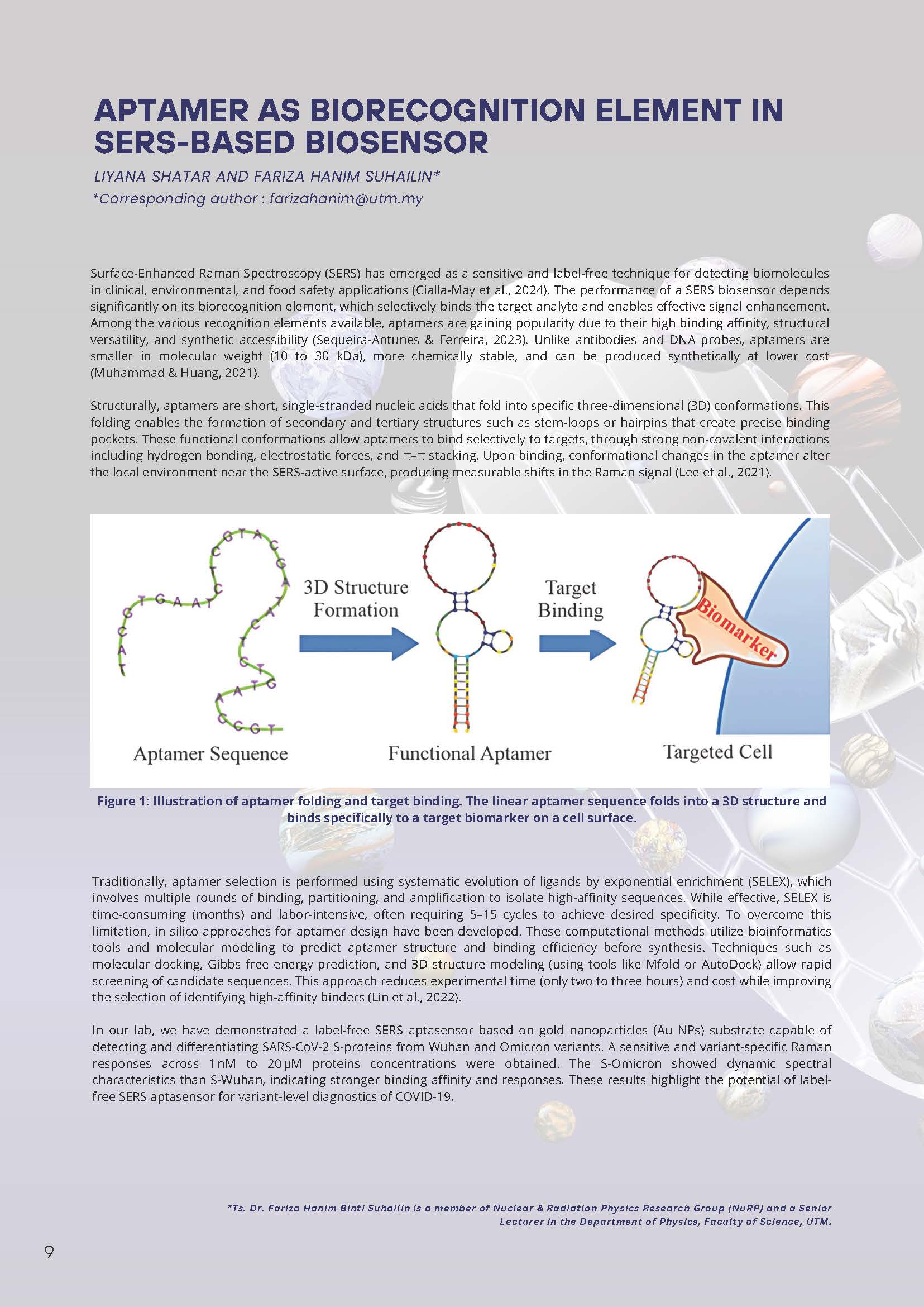
Structurally, aptamers are short, single-stranded nucleic acids that fold into specific three-dimensional (3D) conformations. This folding enables the formation of secondary and tertiary structures such as stem-loops or hairpins that create precise binding pockets. These functional conformations allow aptamers to bind selectively to targets through strong non-covalent interactions, including hydrogen bonding, electrostatic forces, and π–π stacking. Upon binding, conformational changes in the aptamer alter the local environment near the SERS-active surface, producing measurable shifts in the Raman signal (Lee et al., 2021).
Traditionally, aptamer selection is performed using systematic evolution of ligands by exponential enrichment (SELEX), which involves multiple rounds of binding, partitioning, and amplification to isolate high-affinity sequences. While effective, SELEX is time-consuming (months) and labor-intensive, often requiring 5–15 cycles to achieve desired specificity. To overcome this limitation, in silico approaches for aptamer design have been developed. These computational methods utilize bioinformatics tools and molecular modeling to predict aptamer structure and binding efficiency before synthesis. Techniques such as molecular docking, Gibbs free energy prediction, and 3D structure modeling (using tools like Mfold or AutoDock) allow rapid screening of candidate sequences. This approach reduces experimental time (only two to three hours) and cost while improving the selection of identifying high-affinity binders (Lin et al., 2022).
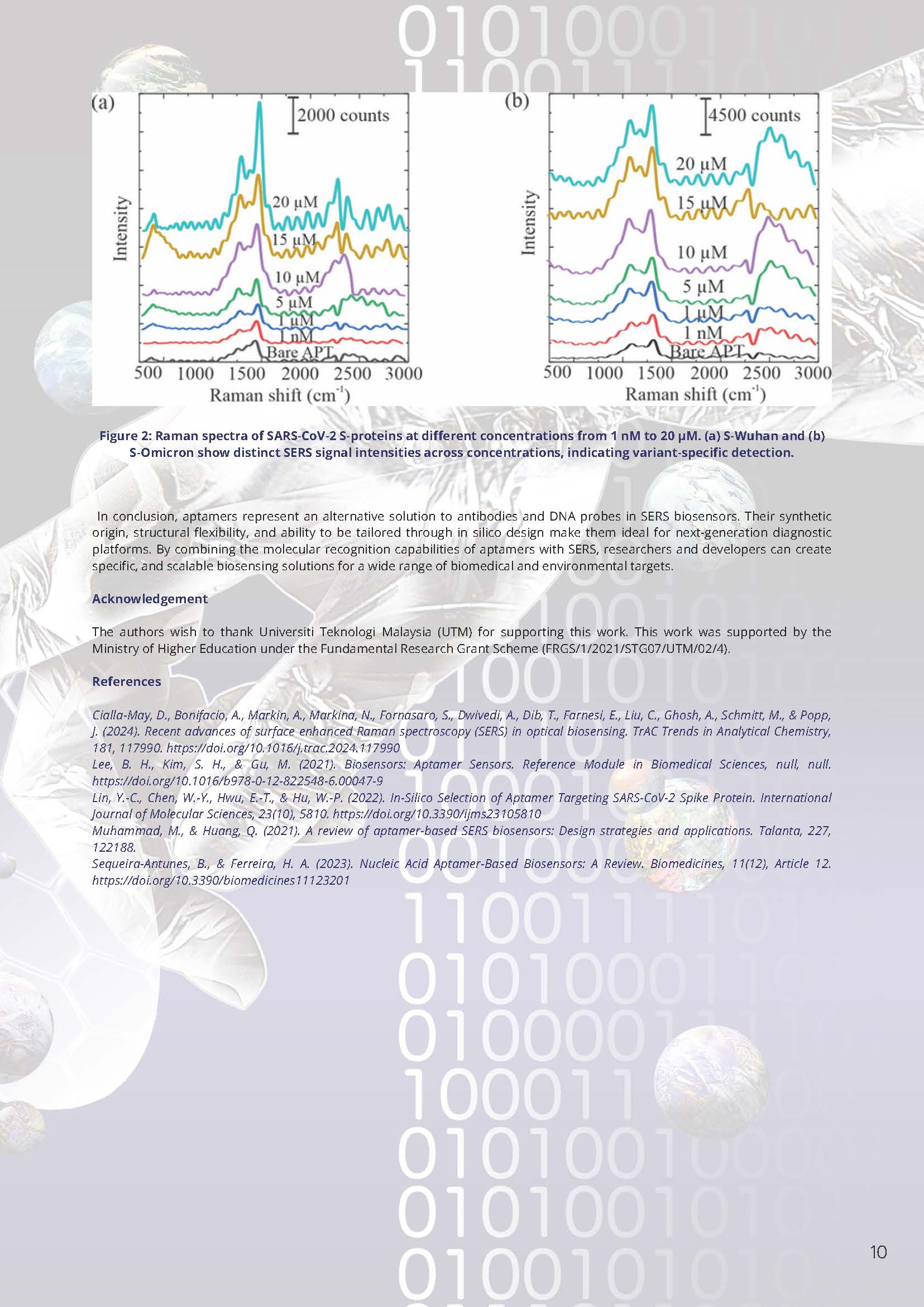
In our lab, we have demonstrated a label-free SERS aptasensor based on gold nanoparticles (Au NPs) substrate capable of detecting and differentiating SARS-CoV-2 S-proteins from Wuhan and Omicron variants. Sensitive and variant-specific Raman responses across 1 nM to 20 µM protein concentrations were obtained. The S-Omicron showed distinct spectral characteristics compared to S-Wuhan, indicating stronger binding affinity and responses. These results highlight the potential of label-free SERS aptasensors for variant-level diagnostics of COVID-19.
Conclusion
In conclusion, aptamers represent an alternative solution to antibodies and DNA probes in SERS biosensors. Their synthetic origin, structural flexibility, and ability to be tailored through in silico design make them ideal for next-generation diagnostic platforms. By combining the molecular recognition capabilities of aptamers with SERS, researchers and developers can create specific and scalable biosensing solutions for a wide range of biomedical and environmental targets.
Acknowledgement
The authors wish to thank Universiti Teknologi Malaysia (UTM) for supporting this work. This work was supported by the Ministry of Higher Education under the Fundamental Research Grant Scheme (FRGS/1/2021/STG07/UTM/02/4).
References
Cialla-May, D., Bonifacio, A., Markin, A., Markina, N., Fornasaro, S., Dwivedi, A., Dib, T., Farnesi, E., Liu, C., Ghosh, A., Schmitt, M., & Popp, J. (2024). Recent advances of surface-enhanced Raman spectroscopy (SERS) in optical biosensing. TrAC Trends in Analytical Chemistry, 181, 117990. https://doi.org/10.1016/j.trac.2024.117990
Lee, B. H., Kim, S. H., & Gu, M. (2021). Biosensors: Aptamer sensors. Reference Module in Biomedical Sciences. https://doi.org/10.1016/B978-0-12-822548-6.00047-9
Lin, Y.-C., Chen, W.-Y., Hwu, E.-T., & Hu, W.-P. (2022). In-silico selection of aptamer targeting SARS-CoV-2 spike protein. International Journal of Molecular Sciences, 23(10), 5810. https://doi.org/10.3390/ijms23105810
Muhammad, M., & Huang, Q. (2021). A review of aptamer-based SERS biosensors: Design strategies and applications. Talanta, 227, 122188.
Sequeira-Antunes, R., & Ferreira, H. A. (2023). Nucleic acid aptamer-based biosensors: A review. Biomedicines, 11(12), Article 12. https://doi.org/10.3390/biomedicines11123201